Abstract
Image texture extraction and analysis are fundamental steps in computer vision. In particular, considering the biomedical field, quantitative imaging methods are increasingly gaining importance because they convey scientifically and clinically relevant information for prediction, prognosis, and treatment response assessment. In this context, radiomic approaches are fostering large-scale studies that can have a significant impact in the clinical practice. In this work, we present a novel method, called CHASM (Cuda, HAralick & SoM), which is accelerated on the graphics processing unit (GPU) for quantitative imaging analyses based on Haralick features and on the self-organizing map (SOM). The Haralick features extraction step relies upon the gray-level co-occurrence matrix, which is computationally burdensome on medical images characterized by a high bit depth. The downstream analyses exploit the SOM with the goal of identifying the underlying clusters of pixels in an unsupervised manner. CHASM is conceived to leverage the parallel computation capabilities of modern GPUs. Analyzing ovarian cancer computed tomography images, CHASM achieved up to \(\sim 19.5\times \) and \(\sim 37\times \) speed-up factors for the Haralick feature extraction and for the SOM execution, respectively, compared to the corresponding C++ coded sequential versions. Such computational results point out the potential of GPUs in the clinical research.
Similar content being viewed by others
1 Introduction
The use of high-performance computing (HPC) is gaining ground in high-dimensional imaging data processing [16], as in the context of hyperspectral image processing [5, 35] and medical image analysis [12]. In particular, for the specific case of medical imaging, along with the acceleration of the training of deep neural networks [47], graphics processing unit (GPU)-powered implementations allowed for real-time performance in image reconstruction [46, 59], segmentation [2], as well as feature extraction [42] and classification [22]. Moreover, multi-core and many-core architectures were exploited to accelerate computationally expensive medical image enhancement and quantification tasks [41, 52, 53].
Feature extraction is the first phase in quantitative imaging as it allows us to perform fundamental tasks in computer vision, such as object detection [55] and representation [48]. Even though deep learning has recently gained ground, conventional machine learning models built on top of handcrafted texture features still play a key role in practical applications, especially relying upon the interpretability of the results [54]. With particular reference to biomedicine, quantitative imaging methods are increasingly gaining importance since they convey scientifically and clinically relevant information for prediction, prognosis, and treatment response assessment [62]. In this context, radiomic approaches are endorsing the transition towards large-scale studies with a relevant impact in the clinical practice [26]. Indeed, radiomics involves the extraction and the analysis of a huge amount of features mined from medical images [25]. The ultimate goal is the objective and quantitative description of tumor phenotypes [13, 26]. Assuming that radiomic features convey information about the different cancer phenotypes, their combination with genomics can enable intra- and inter-tumor heterogeneity studies [45]. Among the radiomic texture feature classes [50], Haralick features are the most well-established and interpretable [18, 19]. These second-order statistics are based on the gray-level co-occurrence matrix (GLCM) that stores the co-occurrence frequency of similar intensity levels over the region (i.e., intensity value pairs). In radiology, Haralick features allow clinicians to assess image regions characterized by heterogeneous/homogeneous areas or local intensity variations [6]. GLCM-based texture features have been extensively exploited in several medical image analysis tasks, such as breast ultrasound (US) classification [15], brain tissue and tumor segmentation on magnetic resonance (MR) images [36, 49], and volume-preserving non-rigid lung computed tomography (CT) image registration [37]. Unfortunately, the computation of these features is considerably burdensome on images characterized by a high bit depth (e.g., 16 bits), such as in the case of medical images that have to convey detailed visual information [31, 43]. As a matter of fact, with the existing computational tools, the range of intensity values of an image must be reduced and limited to achieve an efficient radiomic feature computation [63].
In addition, considering the downstream analyses of the extracted (handcrafted or learned) features, numerous machine learning models can be employed in the context of computer vision [32]. Kohonen self-organizing maps (SOMs) [24] are one of most effective techniques that were applied to biomedical data clustering [3]. A SOM is a special class of artificial neural networks based on the idea of “competitive” learning, able to self-organize the weights in an unsupervised fashion, leading to a spontaneous partitioning of the dataset according to the mutual similarities of the input vectors. SOMs have been used for the analysis of medical images, especially in segmentation tasks in combination with unsupervised clustering [1, 27] or evolutionary computation techniques [36].
Several radiomics toolboxes are available, such as MaZda [51], written in C++, the Computational Environment for Radiological Research (CERR) in MATLAB [4, 10], PyRadiomics in Python [58], and Local Image Feature Extractor (LIFEx) in Java [33]. Importantly, considering 16-bit images, these tools are not suitable for the extraction of the voxel-based feature maps by preserving the initial grayscale range. This limitation is emphasized when dealing with feature extraction tasks on the whole input image, especially for image classification purposes [30].
In this work, we propose a novel GPU-powered pipeline, called CHASM, for the Haralick feature extraction and the downstream unsupervised SOM-based analysis of the feature maps computed on medical images. CHASM exploits HaraliCU [42], a GPU-enabled approach, capable of overcoming the issues of existing tools by effectively computing the feature maps for high-resolution images with their full dynamics of grayscale levels, and CUDA-SOM, a GPU-based implementation of the SOMs for the identification of clusters of pixels in the image. CHASM offloads the computations onto the cores of GPUs, thus allowing us to drastically reduce the running time of the analyses executed on central processing units (CPUs). In the experimental tests performed on ovarian cancer CT images [39], CHASM allowed us to achieved up to \(20\times \) speed-up with respect to the corresponding sequential implementation.
The remainder of the manuscript is organized as follows: Section 2 introduces the basic concepts on Haralick features extraction and on the SOMs. Section 3 describes the proposed GPU-accelerated pipeline, and the obtained results are shown and discussed in Sect. 4. Finally, concluding remarks and future directions are provided in Sect. 5.
2 Background
2.1 Haralick features extraction
Haralick features are GLCM-based texture descriptors that are used to analyze the textural characteristics of an image according to second-order statistics [18, 19]. In medical imaging, these features have shown an appropriate characterization of the cancer imaging phenotype [42]. For instance, the entropy feature is the most promising quantitative imaging biomarker for the analysis of the heterogeneity characterizing cancer imaging [11].
Haralick features are computed from the GLCM, which denotes the co-occurrence frequency of similar intensity levels over the analyzed region. The study conducted in [14] pointed out the existing dependencies among the Haralick features, highlighting how they can be exploited to perform calculations pertaining to other features or intermediate results [42]. Nevertheless, a quantization step (i.e., the compression of the initial intensity range) is generally applied for practical reasons [64], leading to an irreversible loss of information. Even though Brynolfsson et al. stated that the impact of noise is reduced by quantizing the grayscale levels, allowing for obtaining more descriptive Haralick features in MR images, this compression could remarkably affect the discriminative power of the feature-based classification tasks [20]. In any case, the grayscale compression is mostly applied to deal with the computational costs that would be required to calculate these features considering the full grayscale dynamics.
In order to speed up the calculation of Haralick features, HPC solutions can be exploited. For instance, GPUs have been intensively leveraged, being effective computational solutions in life sciences [12, 34]. In the context of Haralick feature extraction on GPUs, different optimization strategies have been presented. For instance, a packed representation of the symmetric GLCM was proposed to only store nonzero elements [14]. By so doing, a simple lookup table, which maps the index of the packed co-matrix, was used to calculate the features reducing the latencies due to memory reads and increasing the overall performances. This efficient implementation allowed for calculating the Haralick features on 12-bit intensity depth images. Another strategy to store the GLCM consists in the meta-GLCM array proposed by Tsai et al. [56], which uses an indirect encoding scheme that fully exploits the GPU memory hierarchy.
The valuable amount of information conveyed by medical images, in terms of both image resolution and pixel depth, should be maintained for automated processing [43], since clinically useful pictorial content could be identified in addition to the naked eye perception. For these motivations, HaraliCU [42] was developed aiming at efficiently keeping the full dynamics of the gray levels (i.e., 16 bits in the case of biomedical images). HaraliCU was tested on brain metastatic tumor MR and ovarian cancer CT images.
2.2 The Self-Organizing Map
The Kohonen SOM [24] is an unsupervised machine learning approach used to perform classification tasks according to the similarity of the data. Technically, a SOM is a class of artificial neural network able to produce low-dimensional (traditionally, bi-dimensional) and discrete representation of the input space.
One important distinction compared to other neural networks is that SOMs exploit a paradigm named competitive learning, which is radically different with respect to classic methods relying upon the minimization of the error by means of gradient descent approaches. Specifically, a SOM is composed of a network of K artificial neurons named units. Usually, the units are all interconnected and logically organized as a \(M \times N\) square or hexagonal grid. Additionally, an input layer composed of D artificial neurons is fully connected to all the units in the SOM, where D is equal to the length of the input samples.
At the beginning of the learning phase, K random weight vectors, \({\mathbf{w }}_{k} \in {\mathbb {R}}^D\), \(k=1, \ldots , K\), are initialized and associated with the units of the network. Then, each input vector \({\mathbf{x }} \in {\mathbb {R}}^D\) in the data set is presented to all units in the SOM. The unit with the most similar weights to the input vector becomes the best matching unit (\({\text {BMU}}\)). In our implementation, we assume a similarity based on an Euclidean distance, i.e.,
Once the \({\text {BMU}}\) is identified, the weights in the network are adjusted toward the input vector using Eq. (2):
where \(\alpha (t)\) is the learning rate at the iteration t. In this work, we used a linearly decreasing learning rate defined as:
with \(\alpha (0)=1\cdot 10^{-1}\) and \(\alpha (t_\text {max})=1\cdot 10^{-3}\). The function \(\varDelta _k(\text {BMU}, t)\) denotes the lateral interaction between the \({\text {BMU}}\) and the unit k during the iteration t. Note that only the units in the set of neighbors of the \({\text {BMU}}\) are updated using Eq. (2). In this work, we exploited the following interaction function:
where \(\sigma (t) = \sigma (0)-(\sigma (0) \frac{t}{t_\text {max}})\). In our implementation, we also used an internal heuristics that calculates the initial \(\sigma \) as:
and sets \(\sigma (t_\text {max})=0\).
Once all weights are updated, the SOM proceeds by analyzing the next sample. The learning algorithm iterates until a stopping criterion is met. In this work, we run the algorithm for \(t_\text {max}=100\) iterations.
Relying upon this peculiar type of learning algorithm—which does not require the samples to be labeled as units that spontaneously self-organize to represent prototypes of the input vectors—SOMs are well-suited for unsupervised learning. As a matter of fact, at the end of the learning process, the samples will be associated with their BMUs in such a way that similar input vectors (with respect to the Euclidean distance) will find place in similar units.
There exist several implementations of SOMs, e.g., the kohonen package for R [61]; the KNLL and SOMpp for C++; the MiniSom and PyCluster libraries for Python [9]. These CPU-based implementations suffer from two main limitations: (1) the computational burden associated with the SOM learning algorithm and (2) the data structures employed, resulting in a very high memory footprint, which increases along with the size of the network.
Various GPU-based implementations of the SOM have been presented in the literature; for instance, in [29], the authors assessed the performance of their GPU version, which parallelizes distance finding, reduction operation to identify the minimum value, and weights adjust, allowing them to speed up the computation up to \(\sim 32\times \) with respect to the CPU. In [8], the authors proposed a parallelization of both the learning and clustering algorithms and applied it to MR image segmentation, achieving up to \(90\times \) speed up with respect to a MATLAB implementation running on the CPU. Unfortunately, both previous works present custom GPU implementations of the SOM, tailored to specific problems. We thus propose here CUDA-SOM as a general-purpose implementation of the SOM accelerated on GPUs using CUDA, freely, and publicly available.
3 The proposed GPU-accelerated method
In this section, we first outline the main CUDA characteristics; then, our GPU implementations of the Haralick feature extraction and SOMs are described in detail. Finally, we present the CHASM framework for medical image analysis.
3.1 CUDA
NVIDIA CUDA is a parallel computing platform and programming model based on many-core streaming multiprocessors (SMs), which adheres to the single instruction multiple data (SIMD) architecture [28]. In CUDA, the CPU (host) offloads the parallel calculations onto one or more GPUs (devices) by using kernels, which are functions launched from the host and replicated so that each GPU thread can run the same code at the same time.
In CUDA, the threads are organized into three-dimensional structures called blocks, which, in turn, compose three-dimensional grids. The CUDA scheduler assigns blocks to the different SMs, which ultimately run them. In each SM, the threads are divided into warps, which are tight groups of 32 threads, executed in locksteps. Considering the CUDA execution pattern, any possible divergent path taken by some threads in a warp should be removed to avoid the serialization of the execution, which would result in a decrease in the overall performance.
CUDA has a complex memory hierarchy divided into multiple memory types, which have their own advantages and drawbacks. For instance, the shared memory is very small but has very low access latency, and it is generally used for intra-block communications. The global memory is large and characterized by high access latency; however, it is visible by all threads and can be used for inter-block communications as well as for communications between the host and the devices. Considering these peculiarities, the data structures should be carefully optimized to reach the theoretical peak performance of the GPU [34].
3.2 HaraliCU
HaraliCU is a GPU-powered tool that realizes an efficient computation of the GLCM and the extraction of an exhaustive set of the Haralick features [42]. The user has granted full control over the settings of HaraliCU, i.e., the distance offset \(\delta \), the orientation \(\theta \), and the window size \(\omega \times \omega \), while the neighborhood \({\mathcal {N}}\) is defined according to \(\delta \) and \(\theta \). In addition, the user can decide the padding conditions (e.g., zero padding or symmetric padding) and the number of quantized gray-level Q.
HaraliCU exploits an effective and efficient encoding, to mitigate the memory requirements related to the allocation of a GLCM having \(2^{16}\) rows and columns for each sliding window, and to the size of each GLCM, which is strictly related to the number of different gray levels inside the considered sliding window. Such an encoding removes all zero elements inside the GLCM and consists in storing each GLCM in a list-based data structure where each element is a pair \(\langle {{\mathsf {GrayPair}}}, {{\mathsf {freq}}} \rangle \), with \({{\mathsf {GrayPair}}}\) being a couple \(\langle i,j \rangle \) of gray levels and \({{\mathsf {freq}}}\) the corresponding frequency of the considered sliding window. Overall, the number of elements composing the GLCM is equal to the number of pairs \(\langle {{\mathsf {reference}}}, {{\mathsf {neighbor}}} \rangle \) that can be identified inside the sliding window, considering the distance \(\delta \) (see [42] for additional information).
The parallelization on the GPU is realized by assigning each pixel of the input image to a thread, since there are no dependencies between the sliding windows. By so doing, each thread computes all features related to its pixel, which is the center of the corresponding window. To fully exploit the GPU acceleration, HaraliCU makes use of a bi-dimensional structure for both the number of blocks and the number of threads. In particular, the number of threads is set to 16 for both the components of the bi-dimensional structure, taking into account the CUDA warp size (i.e., 32 threads) and the limited number of registers, while the number of blocks is set according to the number of the pixels (\(\#\text {pixels}\)) of the input image.
HaraliCU is an open-source software that can be freely downloaded from GitHub at the following address: https://github.com/andrea-tango/HaraliCU. Instructions for the compilation and execution of HaraliCU are provided in the same Web page. HaraliCU requires a NVIDIA GPU along with the version 8 of CUDA (or greater) and the OpenCV library [23] version 3.4.1 (or greater).
3.3 CUDA-SOM
CUDA-SOM is a GPU-based implementation of the self-organizing map [24], where the learning algorithm of the network is parallelized by means of specific kernels that deal with the calculations required to compute the distance between samples and neuron’s weights and to update the network when the \(\text {BMU}\) is identified.
CUDA-SOM supports two GPU-accelerated learning modalities, named online and batch:
-
In the online mode, the weights of the network are updated after each input vector is processed;
-
In the batch mode, the network is updated after the whole training set is analyzed.
Although the batch mode is characterized by a slower convergence compared to online mode, it allows for a higher degree of parallelization, since all calculations of the BMUs for all input vectors can be parallelized across the CUDA cores. For this reason, in this work we exploited the batch mode.
The CUDA kernels implemented in CUDA-SOM allow us to minimize the data transfer between host and device, to the results of the computation, thus reducing the impact of moving data across the PCI-e bus. Moreover, our implementation exploits the Thrust library of CUDA for array scan and reduction, so that the computational time can be further reduced.
CUDA-SOM implements numerous variants of the Kohonen maps. Moreover, even though it integrates several heuristics for the automatic configurations, the user can select a wide array of optional parameters, notably number of neurons; rows and columns of the network; initial and final learning rates; maximum number of iterations of the learning process; radius of the updating function; type of distances used for BMU (e.g., Euclidean, Manhattan, Tanimoto); type of neighbor function (Gaussian, bubble, Mexican hat); type of lattice (square or hexagonal); type of boundary conditions (e.g., toroidal); linear of exponential decay, for both the radius and the learning rate; whether to perform a normalization on the input vectors or not. Moreover, CUDA-SOM gives control on some CUDA-specific settings, e.g., the GPU to be used for the calculations (in the case of multi-GPU systems), or the number of threads per block.
CUDA-SOM is open source and available for downloading on GitHub at the following address: https://github.com/mistrello96/CUDA-SOM.
3.4 CHASM
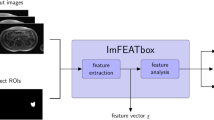
The proposed pipeline begins by extracting the features from the input image by using HaraliCU, which exploits the GPU acceleration. Then, the features of all pixels are transferred on the CPU for further processing. For each pixel, the features are averaged across all directions and linearized. The feature vectors of all pixels are then fed to CUDA-SOM, which performs the unsupervised learning on the GPU. The information about the BMUs for each pixel is returned to the CPU, where it is clustered and mapped onto the original image. The overall functioning of CHASM is schematized in Fig. 1.
In this work, we extract the following 16 Haralick features [18, 19], which are then considered for the unsupervised learning [42]: angular second moment, autocorrelation, cluster prominence, cluster shade, contrast, difference entropy, difference variance, dissimilarity, entropy, homogeneity, inverse difference moment, maximum probability, sum of average, sum of entropy, sum of squares, sum of variance.
The mathematical definitions of the features are provided in Supplementary Materials. Since all the medical images analyzed by CHASM have size \(512\times 512\) pixels, each pixel yields an input vector characterized by \(16 \times 512 \times 512 = 4,194,304\) features.
Examples of results of the unsupervised learning process: a count plot, created according to the number of input vectors assigned to each unit; b corresponding U-matrix; c result of the clustering of the units according to weight distances. The information about which pixels are assigned to each cluster is then mapped onto the original figure
Once the learning process is completed and the BMUs for each input vector are identified, a count plot can be created. In this particular graphical representation, each sample is plotted and assigned to its corresponding BMU. A darker color corresponds to a higher number of samples assigned to the same BMU (see Fig. 2a). Another representation of the outcome of a learning process is the so-called U-matrix, wherein the regions with high inter-neighbor distance are represented with a lighter color. The U-matrix is helpful to provide a visual insight into the boundaries between groups of similar neurons (see Fig. 2b). By using agglomerative clustering, these groups can be identified, and the samples belonging to each unit can be automatically assigned to the proper cluster. The outcome of this process is shown in Fig. 2c. In this work, we exploited the agglomerative clustering implemented in the scikit-learn package, using Euclidean affinity and the Ward linkage criterion [60].
4 Experimental results
As described in the previous section, we validated HaraliCU by comparing the values of the features contrast, correlation, energy, and homogeneity with those extracted using the built-in functions graycomatrix.
4.1 Imaging dataset and tumoral habitats
For the tests presented here, we considered a medical dataset composed of axial contrast-enhanced CT series of patients with high-grade serous ovarian cancer (matrix size: \(512 \times 512\) pixels, pixel spacing: \(\sim 0.65 \times 0.65\,\hbox {mm}^2\), slice thickness: 5.0 mm). All the CT images were encoded in the Digital Imaging and Communications in Medicine (DICOM) format with an intensity depth of 16 bits. Texture features have shown the ability of evaluating intra- and inter-tumor heterogeneity [39, 57]. Pelvic lesions only were selected for this work.
Figure 3 shows two examples of the input CT images along with the corresponding tumoral habitats. In order to simplify the visual interpretation of the results, we used a uniform color coding for the spurious pixels included in disconnected clusters. It is appreciable how CHASM can find patterns to represent both intra-tumoral (Fig. 3a) and inter-tumoral heterogeneity (Fig. 3a) across disconnected lesions.
Examples of CT images with the pelvic lesions outlined by the green contour. The corresponding tumoral habitats, resulting from the unsupervised SOM-based clustering, are overimposed onto the input CT image and displayed at the bottom right of each sub-figure: a tumor composed of a single connected component; b tumor composed of two connected components
4.2 Computational results
The computational performance of the pipeline presented in this work was assessed by independently considering the two steps parallelized on the GPU: Haralick feature extraction and unsupervised SOM-based image pixel clustering.
HaraliCU The CUDA-based version of the Haralick feature extraction, employed in our pipeline, was tested against a CPU version coded in C++, which resulted extremely efficient with respect to the MATLAB version, based on the graycomatrix and graycoprops functions, to extract Haralick features on brain metastasis MR images [42]. As a matter of fact, by varying the grayscale range from \(2^4\) to \(2^9\) levels, we achieved speed-up values around \(50\times \) and \(200\times \), respectively.
The GPU version of HaraliCU was executed on an NVIDIA GeForce GTX Titan X (3072 cores, clock 1.075 GHz, 12 GB of RAM), CUDA toolkit version 8 (driver 387.26), running on a workstation with Ubuntu 16.04 LTS, equipped with a CPU Intel Core i\(7-2600\) (clock 3.4 GHz) and 8 GB of RAM. The CPU version was run on the same workstation, relying upon the computational power provided by the CPU Intel Core i\(7-2600\). The CPU version was compiled by using the GNU C++ compiler (version 5.4.0) with optimization flag -O3, while the GPU version was compiled with the CUDA Toolkit 8 by exploiting the optimization flag -O3 for both CPU and GPU codes.
In order to collect statistically sound results and take into consideration the variability and heterogeneity typically characterizing medical images, we randomly selected 30 images from 3 different patients (10 per patient) affected by brain metastases and 30 images from 3 different patients affected by ovarian cancer. We tested both the CPU and GPU versions by considering various window sizes, that is, \(\omega \in \{3, 7, 11, 15, 19, 23, 27, 31\}\), as well as two different intensity levels (i.e., \(2^8\) and \(2^{16}\)). For each combination of \(\omega \) and intensity levels, we also enabled and disabled the GLCM symmetry to evaluate how the symmetry affects the running time.
The speed-up achieved by HaraliCU considering only \(2^8\) intensity levels increases almost linearly up to \(\omega = 19\) (data not shown, see [42] for details); by disabling the GLCM symmetry and using \(\omega = 31\), we obtained the highest speed-ups of \(12.74\times \) and \(12.71\times \) on brain metastasis (\(256\times 256\) pixels) and ovarian cancer images (\(512\times 512\) pixels), respectively. When the full dynamics of the grayscale levels (i.e., \(2^{16}\)) is considered, HaraliCU outperforms the sequential counterpart, achieving speed-ups up to \(15.80\times \) with \(\omega =31\) and \(19.50\times \) with \(\omega =23\), on brain metastasis and ovarian cancer images, respectively. Taking into account ovarian cancer images, when \(\omega \) is greater than 23 pixels, the speed-up decreases for two reasons. First, since a thread is launched for each pixel, it must consider more neighbor pixels that might have very different gray-level intensities. This corresponds with increasing the required workload that each thread must perform; however, considering that the GPU cores have a lower clock frequency than the CPU cores, the speed-up is clearly reduced. Second, the GPU resources are saturated as the GLCM size associated with each thread may increase due to the high full-dynamic range. In this specific situation, the total GLCM size might overwhelm the capacity of the global memory and some threads might handle different pixels, thus computing the corresponding Haralick features in a sequential way.
CUDA-SOM The performance of CUDA-SOM was assessed by comparing it to a C++ version, running on a single core of the CPU, specifically developed for this work, since the available R implementation of the SOM is limited to a network size of \(150\times 150\) neurons.
We first run a batch of tests to the aim of analyzing the impact of the number of samples and the size of the SOM on the computational time. We employed a machine equipped with 16 GB of RAM, a CPU Intel Core i7 4790k (clock 4.4 Ghz), and an NVIDIA GeForce 1050ti (768 cores, clock 1.392 GHz, 4 GB of RAM).
As reported in Table 1, the running time of the C++ version is lower in the case of small size SOMs (i.e., \(20\times 20\) neurons), while the GPU allows us to reduce the computation time, up to \(5.75\times \), when a SOM having size \(300\times 300\) neurons is trained with 120,000 samples. Additional tests (data not shown) confirmed the trend observed, as the speed-up further increases to \(\sim 7\times \) with a SOM having size \(400\times 400\) neurons.
As a second batch of tests, we compared the performance of different NVIDIA GPUs, i.e., Titan Z (\(2 \times 2880\) cores, clock 0.876 GHz, 6 GB of RAM), Titan X (GM200, 3072 cores, clock 1.075 GHz, 12 GB of RAM), GeForce 1050ti (768 cores, clock 1.392 GHz, 4 GB of RAM), GeForce 1080ti (3584 cores, clock 1582 GHz, 11 GB of RAM), when executing CUDA-SOM with different SOM sizes, considering 60,000 samples and 7 features.
Table 2 reports the speed-up values achieved by each GPU with respect to the C++ implementation. As expected, in the case of small-size SOMs, the CPU was more convenient than the GPUs; moreover, the GeForce 1080ti obtained the best results, by exploiting its highest clock frequency, achieving \(10\times \) speed-up in the case of the SOM with \(400\times 400\) neurons.
Considering the analysis performed on medical images, in the case of ovarian cancer CT, the running time (including file loading) was of 79 and 1020 s in the case of 100 and 1000 iterations, respectively. To understand the advantage of CUDA-SOM, consider that the running time of the same SOM algorithm, implemented with C++ and OpenMP, is 2956 s to complete 100 iterations. This reduction in the running time corresponds to a \(37 \times \) speed-up.
CUDA-SOM was executed on an NVIDIA Tesla P100 (3584 cores, clock 1.329 GHz, 16 GB of RAM), CUDA toolkit version 8 (driver 440.95.01), running on a computer node of the Cambridge Service for Data Driven Discovery (CSD3) with Scientific Linux 7. Each node is equipped with a single CPU Intel Xeon E5-2650 v4 (clock 2.2 GHz), 94 GB of RAM, and up to 4 NVIDIA Tesla P100 GPUs. The CPU version was run on the same node, relying upon the computational power provided by the CPU Intel Xeon E5-2650 v4. The CPU version was compiled by using the GNU C++ compiler (version 5.4.0) with optimization flag -O3, while the GPU version was compiled with the CUDA Toolkit 8.0 by exploiting the optimization flag -O3 for both CPU and GPU codes.
5 Conclusion
Image texture extraction and analysis is playing a key role in quantitative biomedicine, leading to valuable applications in radiomics [13, 25, 26] and radiogenomics [38, 44] research, by also combining heterogeneous sources of information. Therefore, advanced computerized medical image analysis methods, specifically designed to deal with the massive amount of extracted features, as well as to discover intrinsic patterns in the analyzed data, could be beneficial for the definition of imaging biomarkers, which support clinical decision making towards precision medicine [40]. However, these large-scale studies need efficient techniques to drastically reduce the prohibitive running time that is typically required.
In this work, we presented a novel method, named CHASM, which combines two CUDA-based computationally efficient approaches capable of effectively exploiting the power of the modern GPUs: (i) HaraliCU, which is used for Haralick features extraction and allows for accelerating the GLCM computation while keeping the full dynamic range in medical images; (ii) CUDA-SOM, which is exploited for unsupervised image pixel clustering, reduces the running time by leveraging the parallelization of the learning process of the network. Our pipeline was tested on a dataset composed of ovarian cancer CT images. Exploiting the GPU used during the two most computationally demanding phases of the pipeline, we achieved speed-ups up to \(19.50\times \) with HaraliCU and up to \(37\times \) with CUDA-SOM, compared to the CPU version implemented in C++, on our dataset.
As a future development, we plan to improve HaraliCU by exploiting the vectorization of the input image matrices for a better GPU thread block managing. In order to enhance the scalability of the proposed approach, the dynamic parallelism, supported by CUDA, could be exploited to further parallelize the computations as soon as the workload increases (e.g., high window size). Moreover, even though the spatial and temporal locality are already exploited during the GLCM construction process, based on the sliding window, the usage of the GPU memory hierarchy might be optimized [17]. For what concerns CUDA-SOM, the main limitation of the tool is that it currently loads the whole dataset before launching the learning process. Because of that, CUDA-SOM might crash when the dataset exceeds the available GPU memory. We are therefore improving the implementation to read and stream the input vectors during the learning phase, in order to work with datasets of arbitrary size. To further accelerate the learning process, we will also extend CUDA-SOM to leverage low-latency memories (i.e., shared memory and constant memory). Finally, all the computational steps, depicted by the blue dashed blocks in Fig. 1, are currently executed on the CPU and represent a bottleneck of CHASM. We plan to develop them in CUDA to additionally accelerate the whole pipeline.
Considering the biological validation of the texture-derived tumoral habitats [7], the combination of the imaging phenotype and genotype might unravel intra-/inter-tumor heterogeneity, as well as provide valuable insights into treatment response [21, 45], by effectively exploiting advanced computational techniques in oncology [3].
References
Aghajari E, Chandrashekhar GD (2017) Self-organizing map based extended fuzzy c-means (SEEFC) algorithm for image segmentation. Appl Soft Comput 54:347–363. https://doi.org/10.1016/j.asoc.2017.01.003
Al-Ayyoub M, Abu-Dalo AM, Jararweh Y, Jarrah M, Al Sa’d M (2015) A GPU-based implementations of the fuzzy c-means algorithms for medical image segmentation. J Supercomput 71(8):3149–3162. https://doi.org/10.1007/s11227-015-1431-y
Ali HR, Jackson HW, Zanotelli VR, Danenberg E, Fischer JR, Bardwell H et al (2020) Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat Cancer 1(2):163–175. https://doi.org/10.1038/s43018-020-0026-6
Apte AP, Iyer A, Crispin-Ortuzar M, Pandya R, van Dijk LV, Spezi E et al (2018) Extension of CERR for computational radiomics: a comprehensive MATLAB platform for reproducible radiomics research. Med Phys 45(8):3713–3720. https://doi.org/10.1002/mp.13046
Bascoy PG, Quesada-Barriuso P, Heras DB, Argüello F, Demir B, Bruzzone L (2019) Extended attribute profiles on GPU applied to hyperspectral image classification. J Supercomput 75(3):1565–1579. https://doi.org/10.1007/s11227-018-2690-1
Brynolfsson P, Nilsson D, Torheim T, Asklund T, Karlsson CT, Trygg J, Nyholm T, Garpebring A (2017) Haralick texture features from apparent diffusion coefficient (ADC) MRI images depend on imaging and pre-processing parameters. Sci Rep 7(1):4041. https://doi.org/10.1038/s41598-017-04151-4
Cherezov D, Goldgof D, Hall L, Gillies R, Schabath M, Müller H, Depeursinge A (2019) Revealing tumor habitats from texture heterogeneity analysis for classification of lung cancer malignancy and aggressiveness. Sci Rep 9(1):1–9. https://doi.org/10.1038/s41598-019-38831-0
De A, Zhang Y, Guo C (2016) A parallel adaptive segmentation method based on SOM and GPU with application to MRI image processing. Neurocomputing 198:180–189. https://doi.org/10.1016/j.neucom.2015.10.129
De Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20(9):1453–1454. https://doi.org/10.1093/bioinformatics/bth078
Deasy JO, Blanco AI, Clark VH (2003) CERR: a computational environment for radiotherapy research. Med Phys 30(5):979–985. https://doi.org/10.1118/1.1568978
Dercle L, Ammari S, Bateson M, Durand PB, Haspinger E, Massard C, Jaudet C, Varga A, Deutsch E, Soria JC et al (2017) Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Sci Rep 7(1):7952. https://doi.org/10.1038/s41598-017-08310-5
Eklund A, Dufort P, Forsberg D, LaConte SM (2013) Medical image processing on the GPU-past, present and future. Med Image Anal 17(8):1073–1094. https://doi.org/10.1016/j.media.2013.05.008
Gillies RJ, Kinahan PE, Hricak H (2015) Radiomics: images are more than pictures, they are data. Radiology 278(2):563–577. https://doi.org/10.1148/radiol.2015151169
Gipp M, Marcus G, Harder N, Suratanee A, Rohr K, König R, Männer R (2012) Haralick’s texture features computation accelerated by GPUs for biological applications. Modeling simulation and optimization of complex processes. Springer, Berlin, pp 127–137. https://doi.org/10.1007/978-3-642-25707-011
Gómez W, Pereira W, Infantosi AFC (2012) Analysis of co-occurrence texture statistics as a function of gray-level quantization for classifying breast ultrasound. IEEE Trans Med Imag 31(10):1889–1899. https://doi.org/10.1109/TMI.2012.2206398
Gulo CA, Sementille AC, Tavares JMR (2019) Techniques of medical image processing and analysis accelerated by high-performance computing: a systematic literature review. J Real Time Image Process. https://doi.org/10.1007/s11554-017-0734-z
Gupta S, Xiang P, Zhou H (2013) Analyzing locality of memory references in GPU architectures. In: Proceedings of ACM SIGPLAN Workshop on Memory Systems Performance and Correctness. ACM, p 12. https://doi.org/10.1145/2492408.2492423
Haralick RM (1979) Statistical and structural approaches to texture. Proc IEEE 67(5):786–804. https://doi.org/10.1109/PROC.1979.11328
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC–3(6):610–621. https://doi.org/10.1109/TSMC.1973.4309314
Jen CC, Yu SS (2015) Automatic detection of abnormal mammograms in mammographic images. Expert Syst Appl 42(6):3048–3055. https://doi.org/10.1016/j.eswa.2014.11.061
Jiménez-Sánchez A, Cybulska P, Mager KL, Koplev S, Cast O, Couturier DL et al (2020) Unraveling tumor-immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Genet Nat. https://doi.org/10.1038/s41588-020-0630-5
Junior JRF, Oliveira MC, de Azevedo-Marques PM (2017) Integrating 3D image descriptors of margin sharpness and texture on a GPU-optimized similar pulmonary nodule retrieval engine. J Supercomput 73(8):3451–3467. https://doi.org/10.1007/s11227-016-1818-4
Kaehler A, Bradski G (2016) Learning OpenCV 3: computer vision in C++ with the OpenCV library, vol 1. O’Reilly Media, Inc, Sebastopol
Kohonen T (1990) The self-organizing map. Proc IEEE 78(9):1464–1480. https://doi.org/10.1109/5.58325
Lambin P, Leijenaar RT, Deist TM, Peerlings J, de Jong EE, van Timmeren J et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749. https://doi.org/10.1038/nrclinonc.2017.141
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48(4):441–446. https://doi.org/10.1016/j.ejca.2011.11.036
Logeswari T, Karnan M (2010) Hybrid self organizing map for improved implementation of brain MRI segmentation. In: Proceedings of International Conference on Signal Acquisition and Processing. IEEE, pp 248–252. https://doi.org/10.1109/ICSAP.2010.56
Luebke D (2008) CUDA: scalable parallel programming for high-performance scientific computing. In: Proceedings 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI). IEEE, pp 836–838. https://doi.org/10.1109/ISBI.2008.4541126
McConnell S, Sturgeon R, Henry G, Mayne A, Hurley R (2012) Scalability of self-organizing maps on a GPU cluster using OpenCL and CUDA. J Phys Conf Ser 341:012018. https://doi.org/10.1088/1742-6596/341/1/012018
Militello C, Rundo L, Minafra L, Cammarata FP, Calvaruso M, Conti V, Russo G (2020) MF2C3: Multi-feature fuzzy clustering to enhance cell colony detection in automated clonogenic assay evaluation. Symmetry 12(5):773. https://doi.org/10.3390/sym12050773
Militello C, Vitabile S, Rundo L, Russo G, Midiri M, Gilardi MC (2015) A fully automatic 2D segmentation method for uterine fibroid in MRgFUS treatment evaluation. Comput Biol Med 62:277–292. https://doi.org/10.1016/j.compbiomed.2015.04.030
Nanni L, Ghidoni S, Brahnam S (2017) Handcrafted versus non-handcrafted features for computer vision classification. Pattern Recogn 71:158–172. https://doi.org/10.1016/j.patcog.2017.05.025
Nioche C, Orlhac F, Boughdad S, Reuzé S, Goya-Outi J, Robert C, Pellot-Barakat C, Soussan M, Frouin F, Buvat I (2018) LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res 78(16):4786–4789. https://doi.org/10.1158/0008-5472.CAN-18-0125
Nobile MS, Cazzaniga P, Tangherloni A, Besozzi D (2016) Graphics processing units in bioinformatics, computational biology and systems biology. Brief Bioinform 18(5):870–885. https://doi.org/10.1093/bib/bbw058
Ordóñez Á, Argüello F, Heras DB, Demir B (2020) GPU-accelerated registration of hyperspectral images using KAZE features. J Supercomput. https://doi.org/10.1007/s11227-020-03214-0
Ortiz A, Górriz J, Ramírez J, Salas-Gonzalez D, Llamas-Elvira JM (2013) Two fully-unsupervised methods for MR brain image segmentation using SOM-based strategies. Appl Soft Comput 13(5):2668–2682. https://doi.org/10.1016/j.asoc.2012.11.020
Park S, Kim B, Lee J, Goo JM, Shin YG (2011) GGO nodule volume-preserving nonrigid lung registration using GLCM texture analysis. IEEE Trans Biomed Eng 58(10):2885–2894. https://doi.org/10.1109/TBME.2011.2162330
Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, Hricak H, Sutton EJ, Morris EA (2018) Background, current role, and potential applications of radiogenomics. J Magn Reson Imag 47(3):604–620. https://doi.org/10.1002/jmri.25870
Rundo L, Beer L, Ursprung S, Martin-Gonzalez P, Markowetz F, Brenton JD, Crispin-Ortuzar M, Sala E, Woitek R (2020) Tissue-specific and interpretable sub-segmentation of whole tumour burden on CT images by unsupervised fuzzy clustering. Comput Biol Med. https://doi.org/10.1016/j.compbiomed.2020.103751
Rundo L, Pirrone R, Vitabile S, Sala E, Gambino O (2020) Recent advances of HCI in decision-making tasks for optimized clinical workflows and precision medicine. J Biomed Inform 108:103479. https://doi.org/10.1016/j.jbi.2020.103479
Rundo L, Tangherloni A, Cazzaniga P, Nobile MS, Russo G, Gilardi MC et al (2019) A novel framework for MR image segmentation and quantification by using MedGA. Comput Methods Progr Biomed 176:159–172. https://doi.org/10.1016/j.cmpb.2019.04.016
Rundo L, Tangherloni A, Galimberti S, Cazzaniga P, Woitek R, Sala E, et al. (2019) HaraliCU: GPU-powered Haralick feature extraction on medical images exploiting the full dynamics of gray-scale levels. In: Malyshkin V (ed) Proceedings of International Conference on Parallel Computing Technologies (PaCT), LNCS, vol 11657. Springer International Publishing, Cham, Switzerland, pp 304–318. 978-3-030-25636-4\_24
Rundo L, Tangherloni A, Nobile MS, Militello C, Besozzi D, Mauri G, Cazzaniga P (2019) MedGA: a novel evolutionary method for image enhancement in medical imaging systems. Expert Syst Appl 119:387–399. https://doi.org/10.1016/j.eswa.2018.11.013
Rutman AM, Kuo MD (2009) Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 70(2):232–241. https://doi.org/10.1016/j.ejrad.2009.01.050
Sala E, Mema E, Himoto Y, Veeraraghavan H, Brenton JD, Snyder A, Weigelt B, Vargas HA (2017) Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol 72(1):3–10. https://doi.org/10.1016/j.crad.2016.09.013
Schellmann M, Gorlatch S, Meiländer D, Kösters T, Schäfers K, Wübbeling F, Burger M (2011) Parallel medical image reconstruction: from graphics processing units (GPU) to grids. J Supercomput 57(2):151–160. https://doi.org/10.1007/s11227-010-0397-z
Shen D, Wu G, Suk HI (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
Soh LK, Tsatsoulis C (1999) Texture analysis of SAR sea ice imagery using gray level co-occurrence matrices. IEEE Trans Geosci Remote Sens 37(2):780–795. https://doi.org/10.1109/36.752194
Sompong C, Wongthanavasu S (2017) An efficient brain tumor segmentation based on cellular automata and improved tumor-cut algorithm. Expert Syst Appl 72:231–244. https://doi.org/10.1016/j.eswa.2016.10.064
Stoyanova R, Takhar M, Tschudi Y, Ford JC, Solórzano G, Erho N, Balagurunathan Y, Punnen S, Davicioni E, Gillies RJ et al (2016) Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res 5(4):432. https://doi.org/10.21037/tcr.2016.06.20
Szczypiński PM, Strzelecki M, Materka A, Klepaczko A (2009) MaZda-a software package for image texture analysis. Comput Methods Progr Biomed 94(1):66–76. https://doi.org/10.1016/j.cmpb.2008.08.005
Tangherloni A, Spolaor S, Cazzaniga P, Besozzi D, Rundo L, Mauri G, Nobile MS (2019) Biochemical parameter estimation vs. benchmark functions: a comparative study of optimization performance and representation design. Appl Soft Comput 81:105494. https://doi.org/10.1016/j.asoc.2019.105494
Tangherloni A, Spolaor S, Rundo L, Nobile MS, Cazzaniga P, Mauri G, Liò P, Merelli I, Besozzi D (2019) GenHap: a novel computational method based on genetic algorithms for haplotype assembly. BMC Bioinform 20:172. https://doi.org/10.1186/s12859-019-2691-y
Torheim T, Malinen E, Kvaal K, Lyng H, Indahl UG, Andersen EK, Futsæther CM (2014) Classification of dynamic contrast enhanced MR images of cervical cancers using texture analysis and support vector machines. IEEE Trans Med Imag 33(8):1648–1656. https://doi.org/10.1109/TMI.2014.2321024
Trivedi MM, Harlow CA, Conners RW, Goh S (1984) Object detection based on gray level cooccurrence. Comput Vis Graph Image Process 28(2):199–219. https://doi.org/10.1016/S0734-189X(84)80022-5
Tsai HY, Zhang H, Hung CL, Min G (2017) GPU-accelerated features extraction from magnetic resonance images. IEEE Access 5:22634–22646. https://doi.org/10.1109/ACCESS.2017.2756624
Vargas HA, Veeraraghavan H, Micco M, Nougaret S, Lakhman Y, Meier AA, Sosa R, Soslow RA, Levine DA, Weigelt B et al (2017) A novel representation of inter-site tumour heterogeneity from pre-treatment computed tomography textures classifies ovarian cancers by clinical outcome. Eur Radiol 27(9):3991–4001. https://doi.org/10.1007/s00330-017-4779-y
van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillion-Robin JC, Pieper S, Aerts HJ (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Vishnevskiy V, Walheim J, Kozerke S (2020) Deep variational network for rapid 4D flow MRI reconstruction. Nat Mach Intell 2(4):228–235. https://doi.org/10.1038/s42256-020-0165-6
Ward JH Jr (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58(301):236–244. https://doi.org/10.1080/01621459.1963.10500845
Wehrens R, Buydens LM et al (2007) Self- and super-organizing maps in R: the Kohonen package. J Stat Softw 21(5):1–19. https://doi.org/10.18637/jss.v021.i05
Yankeelov TE, Mankoff DA, Schwartz LH, Lieberman FS, Buatti JM, Mountz JM, Erickson BJ, Fennessy FM, Huang W, Kalpathy-Cramer J et al (2016) Quantitative imaging in cancer clinical trials. Clin Cancer Res 22(2):284–290. https://doi.org/10.1158/1078-0432.CCR-14-3336
Yip SS, Aerts HJ (2016) Applications and limitations of radiomics. Phys Med Biol 61(13):R150. https://doi.org/10.1088/0031-9155/61/13/R150
Zwanenburg A, Vallières M, Abdalah MA, Aerts HJ, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295(2):328–338. https://doi.org/10.1148/radiol.2020191145
Acknowledgements
This work was partially supported by The Mark Foundation for Cancer Research and Cancer Research UK Cambridge Centre [C9685/A25177]. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. This work was performed using resources provided by the Cambridge Service for Data Driven Discovery (CSD3) operated by the University of Cambridge Research Computing Service (www.csd3.cam.ac.uk), provided by Dell EMC and Intel using Tier-2 funding from the Engineering and Physical Sciences Research Council (capital Grant EP/P020259/1), and DiRAC funding from the Science and Technology Facilities Council (www.dirac.ac.uk).
Funding
Open Access funding provided by Università degli Studi di Milano - Bicocca.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rundo, L., Tangherloni, A., Cazzaniga, P. et al. A CUDA-powered method for the feature extraction and unsupervised analysis of medical images. J Supercomput 77, 8514–8531 (2021). https://doi.org/10.1007/s11227-020-03565-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-020-03565-8