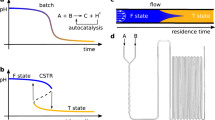
Abstract
Autocatalytic cycles are rather widespread in nature and in several theoretical models of catalytic reaction networks their emergence is hypothesized to be inevitable when the network is or becomes sufficiently complex. Nevertheless, the emergence of autocatalytic cycles has been never observed in wet laboratory experiments. Here, we present a novel model of catalytic reaction networks with the explicit goal of filling the gap between theoretical predictions and experimental findings. The model is based on previous study of Kauffman, with new features in the introduction of a stochastic algorithm to describe the dynamics and in the possibility to increase the number of elements and reactions according to the dynamical evolution of the system. Furthermore, the introduction of a temporal threshold allows the detection of cycles even in our context of a stochastic model with asynchronous update. In this study, we describe the model and present results concerning the effect on the overall dynamics of varying (a) the average residence time of the elements in the reactor, (b) both the composition of the firing disk and the concentration of the molecules belonging to it, (c) the composition of the incoming flux.





Similar content being viewed by others
Notes
We also mention the lipid-world hypothesis (Segre et al. 1998).
The reaction volume was taken to be similar to that of a small bacterium, i.e. 1 μm3.
In this case one single reaction is enough to produce two different polymers of equal length, so the total amount of reactions have to be reduced by 2 × 22L−L = 2L+1 reactions.
Notice also that in all the simulations that we describe in this article we use firing disks and incoming fluxes without any ACS and that in all the simulations the alphabet is composed of two letters, A and B.
References
Bagley RJ, Farmer JD (1991) Spontaneous emergence of a metabolism. Artif Life II:93–140. Santa Fe Institute Studies in the Sciences of Complexity, X
Bower JM, Bolouri H (2004) Computational modeling of genetic and biochemical networks. Computational molecular biology. The MIT Press, Cambridge
Carletti T, Serra R, Poli I, Villani M, Filisetti A (2008) Sufficient conditions for emergent synchronization in protocell models. J Theor Biol 254(4):741–751
Dyson FJ (1985) Origins of life. Cambridge University Press, Cambridge
Eigen M, Schuster P (1977) The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Die Naturwissenschaften 11(64):541–565
Eigen M, Shuster P (1979) The hypercycle: a principle of natural self-organization. Springer, Berlin
Farmer JD, Kauffman SA, Packard NH (1986) Autocatalytic replication of polymers. Physica D 22(2):50–67
Filisetti A, Serra R, Carletti T, Villani M, Poli I (2008) Synchronization phenomena in protocell models. Biophys Rev Lett 3(1/2):325–342
Filisetti A, Graudenzi A, Serra R, Villani M, De Lucrezia D, Fuchslin RM, Kauffman SA, Packard N, Poli I (2011) A stochastic model of the emergence of autocatalytic cycles. J Syst Chem 11:2. doi:10.1186/1759-2208-2-2
Gillespie DT (1977) Exact stochastic simulation of coupled chemical reactions. J Phys Chem 81(25):2340–2361
Gillespie DT (2007) Stochastic simulation of chemical kinetics. Annu Rev Phys Chem 58(1):35–55
Jain S, Krishna S (2001) A model for the emergence of cooperation, interdependence, and structure in evolving networks. Proc Natl Acad Sci USA 98(2):543–547
Kaneko K (2006) Life: an introduction to complex systems biology. Understanding complex systems. Springer, New York
Kauffman SA (1986) Autocatalytic sets of proteins. J Theor Biol 119(1):1–24
Kauffman SA (2008) Reinventing the sacred: a new view of science, reason and religion. Basic Books, New York
Lutkepohl H (1996) Handbook of matrices. Wiley, New York, xvi + 304 pp, ISBN 0-471-96688-6
Munteanu A, Attolini C, Rasmussen S, Ziock H, Solé R (2006) Generic Darwinian selection in protocell assemblies. SFI-WP 06-09-032, SFI Working Papers, Santa Fe Institute
Rasmussen S, Chen L, Deamer D, Krakauer DC, Packard NH, Stadler PF, Bedau MA (2004) Transitions from nonliving to living matter. Science 303:963–965
Segre D, Lancet D, Kedem O, Pilpel Y (1998) Graded autocatalysis replication domain (GARD): kinetic analysis of self-replication in mutually catalytic sets. Orig Life Evol Biosph 28(4–6):501–514
Serra R, Carletti T, Poli I (2007) Synchronization phenomena in surface-reaction models of protocells. Artif Life 13:1–16
Szostak JW, Bartel DP, Luisi PL (2001) Synthesizing life. Nature 409(6818):387–390
Acknowledgments
This study has been partially supported by the Fondazione di Venezia, http://www.fondazionedivenezia.it (DICE project). Interesting discussions with Davide De Lucrezia and Timoteo Carletti are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Filisetti, A., Graudenzi, A., Serra, R. et al. A stochastic model of autocatalytic reaction networks. Theory Biosci. 131, 85–93 (2012). https://doi.org/10.1007/s12064-011-0136-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12064-011-0136-x